The article with participation of NIOCh's researchers is published in
Computational and Structural Biotechnology Journal (IF 7,271) :
Two alternative conformations of mRNA in the human ribosome during elongation and termination of translation as revealed by EPR spectroscopy
Abstract
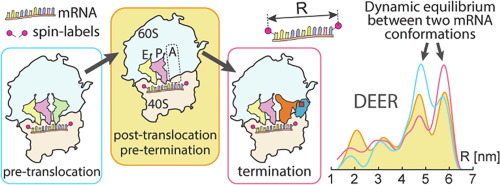
The conformation of mRNA in the region of the human 80S ribosome decoding site was monitored using 11-mer mRNA analogues that bore nitroxide spin labels attached to the terminal nucleotide bases. Intramolecular spin–spin distances were measured by DEER/PELDOR spectroscopy in model complexes mimicking different states of the 80S ribosome during elongation and termination of translation. The measurements revealed that in all studied complexes, mRNA exists in two alternative conformations, whose ratios are different in post-translocation, pre-translocation and termination complexes. We found that the presence of a tRNA molecule at the ribosomal A site decreases the relative share of the more extended mRNA conformation, whereas the binding of eRF1 (alone or in a complex with eRF3) results in the opposite effect. In the termination complexes, the ratios of mRNA conformations are practically the same, indicating that a part of mRNA bound in the ribosome channel does not undergo significant structural alterations in the course of completion of the translation. Our results contribute to the understanding of mRNA molecular dynamics in the mammalian ribosome channel during translation.
Альметрики:


